By: Kirk E. Anderson

Figure 1. Duan Copeland, Ph.D., inspecting hives in Tucson, AZ.
Researchers in Tucson, Arizona, are developing an AI-powered application to simplify and automate honey bee brood disease diagnosis and promote effective disease management. Leading the charge is Dr. Duan Copeland, a postdoctoral researcher in Dr. Kirk E. Anderson’s Lab, dedicated to diagnosing brood disease using only a smartphone photograph. “What can be challenging for even master beekeepers are the subtle visual cues produced by various disease-causing agents that target honey bee larvae, including bacteria, viruses and fungi” said Copeland. “We can train an AI program to recognize these differences in the same way an expert beekeeper or apiary inspector could.”
Connecting the dots
This brood disease research was sparked by Illinois State Apiary Inspector Jim Wellwood. In 2015, Jim came to visit the Tucson lab in conjunction with the annual apiary inspectors meeting. I was intrigued by our conversation, so I began a study of brood disease sampling throughout Illinois. Using these findings as preliminary data, the Anderson lab was awarded a large NIFA grant that included disease experts and co-PIs, Dr. Jay Evans and Dr. Meghan Milbrath, titled: Using big data to improve diagnosis of larval disease in honey bees. We began this project by high-throughput sequencing the bacterial microbiomes of third, fourth and fifth instar larvae to document disease progression across six diseased and one disease-free apiary. Simultaneously, we photographed the same larvae at high resolution (https://www.nature.com/articles/s41598-023-28085-2).
Of the apiaries we selected for deep sequencing, five of seven were experiencing EFB symptoms, one was asymptomatic and one had “melty” symptoms. Our approach sampling throughout larval development showed that EFB disease can manifest in a variety of ways. Similarly, recent results sequencing the genomes of EFB causative agent, M. plutonius, indicate that different bacterial strains have radically different personalities, and differ substantially in their ability to cause disease. The behavior of M. plutonius as a larval commensal, opportunist or pathogen is defined by a collection of virulence genes that allow it to exploit larvae. Additionally, there are genes in the M. plutonius genome that confer survival in the worker gut and hive environments, including royal jelly and honey.
Surprisingly, asymptomatic larval microbiomes frequently contained M. plutonius, including those sampled from asymptomatic apiaries and colonies. Some of this result came from the Tucson Lab Apiary where we rarely, if ever, experience EFB symptoms, yet a significant proportion (41%, 31 of 75) of the healthy larval microbiomes contained M. plutonius. Similarly, at one of the Illinois apiaries with no EFB symptoms, 75% (18 of 24) of asymptomatic larvae were positive for M. plutonius. The high-throughput method used to acquire this data is more sensitive than most tests and detects small amounts of bacteria providing a more complete picture of “who” is in there versus traditional culturing methods.
We discovered a variety of bacteria that occur as relatively harmless commensals in first through third instar larvae. As demonstrated for other species, these molecular patterns serve to “train” the immune system of developing larvae. We found that the progression of European Foulbrood (EFB) differed significantly by apiary due to secondary invaders and differences in beneficial bacteria. The discovered secondary invaders were very different from those identified with earlier culture-based methods with the single exception of Enterococcus faecalis, common to all EFB diseased apiaries. In fact, the presence and abundance of E. faecalis was positively associated with that of M. plutonius across multiple apiaries especially in asymptomatic larvae. This pattern of association is consistent with culture-based results and often indicates synergy among bacterial species. This pattern wasn’t limited to E. faecalis. A number of bacteria that commonly occur throughout the honey bee social network revealed their opportunistic nature, increasing with EFB disease progression. These species were Frischella perrara, the bacterium that commonly forms a scab in the worker gut where the host waste products are excreted, Apilactobacillus kunkeei an extremophilic bacterium that specializes on honey, and Fructobacillus fructosus, a lesser honey specialist that also occurs in healthy larvae but is scarce in the adult gut.
At one of the apiaries, one colony showed symptoms of Varroosis (Parasitic mite syndrome: PMS, also known as Idiopathic Brood Disease Syndrome: IBDS). Digging a little deeper, we found that Acute Bee Paralysis Virus (ABPV) levels were extraordinarily high in the symptomatic larvae, and correlated absolutely with the “melty, deflated and sunken” symptomology recorded by the apiary inspector. Critically, all of these comprehensively defined disease states and larval stages were recorded with high-resolution digital imaging. After many weeks examining these photos and disease states, we hypothesized that larval symptoms alone could be leveraged to accurately diagnose disease.
While it is known that paralytic viruses can infect larvae, honey bee science lacks an understanding of the microbes that either cause or result from “EFB-like” brood disease, molten brood, melty larvae, Parasitic mite syndrome and Varroosis. As part of our recent NIFA grant, we have been funded to unravel this can of worms; the crud, snot brood, melty brood, and mysteries that surround EFB-like brood disease. Our approach first relied on verbal descriptors, which we quickly determined are overly subjective, and woefully inadequate for diagnosis. We instead opted for the “picture does not lie” approach. In other words, a picture is comprised of pixels of quantifiable brightness, color and hue, arranged to form emergent properties that the computer program (or your mind) has been trained to interpret as shapes, in this case larvae, healthy or diseased.
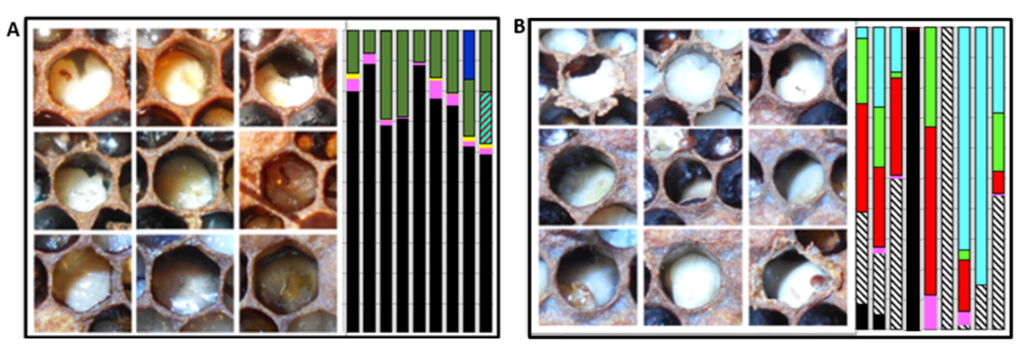
Figure 2. Images of larvae and their associated bacterial microbiomes (vertical bars) from two different apiaries with brood disease: A) Larvae infected with EFB (black) and secondary invader Fructobacillus fructosus (dark green). B) Larvae infected with Acute Bee Paralysis Virus (ABPV) and associated opportunistic bacteria that carry antibiotic resistance genes including Serratia marcescens (red) and Frischella perrara (diagonal striped). The beekeeper treated apiary “B” with antibiotics, but the treatment was ineffective.
Traditional methods used to diagnose brood disease in the field require years of expertise. Like the rest of us novice beekeepers, perhaps it required a few seasons of dedicated beekeeping to form a reliable picture of the major honey bee brood diseases; conditions like chalkbrood, sacbrood, AFB, EFB, and EFB-like. Misdiagnosis is common, and prompts the unnecessary use of antibiotics, which disrupts the balance of the honey bee’s native microbiome. In turn, this disruption promotes the emergence of antibiotic-resistant strains of bacteria, further complicating disease management. In the case of the ABPV apiary (Fig. 2), the beekeeper assumed EFB disease and applied antibiotics. As illuminated by our metagenomic analysis, the antibiotics depleted the larval microbiome of beneficial bacteria and likely contributed to ABPV disease progression by removing the native barrier to opportunistic disease.
The future with AI
By leveraging the power of AI, the team aims to minimize the risk of misdiagnosis, reduce the reliance on antibiotics and ultimately contribute to more effective disease management practices for honey bees. Artificial Intelligence has already demonstrated success in honey bee research. Examples of image-based AI include identifying subspecies by wing patterns, detecting parasitic mites, tracking pollen foraging behavior and identification of comb resources.

Figure 3. Duan Copeland evaluating the efficiency of prototype AI models.
“Even a novice could look at diseased larvae and tell you some physical characteristics about it… this one looks yellow, this one is brown, this one’s melty,” he says. “An AI program can pick up on these same patterns, but it’s crucial we have the correct diagnosis and labeling to facilitate the AI training process.” Anderson’s team is collaborating with the Bee Disease Diagnostic Service in Beltsville, Maryland and apiary inspectors around the U.S. to expand their AI image training dataset by including a wider variety of AFB, EFB, viral and fungal disease. This partnership has broadened our collection of digital images and diverse brood disease phenotypes. In addition, diseased brood samples undergo molecular diagnostic screening and microbiome analysis in Anderson’s lab to ensure the correct diagnosis. By incorporating a more comprehensive range of disease symptoms, the researchers aim to continually update and improve the AI application’s diagnostic capabilities as a honey bee health management tool. “The honey bee pathosphere is constantly evolving, so we want to keep our AI database up to date with what is being seen out there,” Copeland says.
AI in the palm of your hands
The culmination of this research will be the development of a digital product, the Big Data Brood Disease (BDBD) app. This technology will assist the broader beekeeping community by substantially increasing the probability of an accurate diagnosis when the symptoms are unclear, or when a beekeeper has little experience and has not yet formed a reliable picture of various larval disease states. Following the recent big boom in hobbyist and beginner beekeepers across the nation, our tool will significantly reduce the development of antibiotic resistance in both pathogenic, opportunistic and beneficial bacteria. The early and rapid identification of disease outbreaks will facilitate the decision to apply antibiotics or alternative approved treatments. As a result, the BDBD app will contribute significantly to healthier bee populations and more sustainable beekeeping practices. If you are dealing with larval disease outbreaks, and would like to contribute to this project, please contact the Anderson lab NIFA project manager Brendon Mott: Brendon.Mott@usda.gov.







